Illustrative GSK DPhil projects from related programmes
[Offered to students on the ILESLA Programme in 2025]
Supervisors
- Robin Thompson (Maths)
- Will Hart (Maths)
- Rajat Desikan (GSK)
- Anna Sher (GSK)
Infectious diseases are responsible for substantial morbidity and mortality worldwide. For many viruses, vaccination is a key intervention, reducing the likelihood of infection and limiting the negative consequences of breakthrough infections. However, optimising vaccine development and deployment is complex. Key challenges include determining the optimal vaccine dose and prime-boost intervals, as well as deciding which individuals within a population should be vaccinated first.
Mathematical modelling can be used to inform these decisions and improve vaccination efficacy.
This project focuses on optimising vaccine dosing. While higher doses typically provide greater protection, they are more costly and may be associated with increased risks of negative side effects or reduced immunogenicity, depending on the dose–response relationship. Currently, vaccine doses are generally selected to be the lowest possible dose that still produces a sufficiently strong immune response in clinical trial participants.
The core aim of this project is to develop a multi-scale mathematical modelling framework to test whether this within-host dose optimisation strategy is truly optimal at the population level. Specifically, the project will investigate whether a slightly higher dose could result in greater overall population benefit (e.g. fewer cases) that outweighs the costs associated with increased side effects.
Methods and Skills
Key mathematical approaches used in the full DPhil project include:
- Stochastic and deterministic transmission modelling
- Bayesian parameter inference
- Numerical solution of ordinary differential equations (ODEs)
The project also involves computer programming in a language of the student’s choice. Members of Prof. Thompson’s group currently use Python, MATLAB, R, or Julia.
Opportunities for Extension
While the initial rotation project focuses on developing the modelling framework, there are many opportunities to extend this work into a three-year DPhil project. For example:
- Applying the framework to vaccination data for multiple pathogens
- Exploring vaccine dosing strategies in populations with different characteristics and vulnerabilities, such as:
- Paediatric populations
- General adult populations
- Older adult populations
[Project chosen by Abbie Evans, SABS R3 Programme 2023]
Supervisors
- Robin Thompson (Maths)
- Will Hart (Maths)
Infectious diseases are responsible for substantial morbidity and mortality around the world. However, despite the global nature of the challenge of limiting the negative impacts of disease, population-scale transmission is influenced by pathogen dynamics arising at the individual-host level. The potential for an infected individual to infect others is linked to their pathogen load, as well as other factors such as their behaviour. Severe infections can have immediate negative consequences, such as host death, and there is accumulating evidence that even infections that do not immediately have adverse outcomes can lead to subsequent complications. It has been suggested that interventions such as vaccination may therefore reduce the risk of later complications, with recent evidence indicating that Shingrix vaccination (to prevent shingles) may be associated with a reduced risk of dementia several years later.
This project focuses on viral infections. The core aim is to develop mathematical and computational models to explore the impacts of within-host viral dynamics on population-scale transmission and negative outcomes in host populations. Models will be built to characterise the within-host dynamics of a range of viruses, including Ebolavirus and Epstein-Barr virus. Key questions that the models will be used to answer include, among others: i) Which pharmaceutical and non-pharmaceutical interventions can be used to reduce the risk of large Ebola outbreaks?; ii) Which features of within-host dynamics could be responsible for the association between Epstein-Barr virus infections and subsequent onset of neurological disease?
In addition to construction of the underlying models and modelling methods, the project will involve the development of software based on the models that can be used by other scientists and public health policy advisors. Potential uses include the design of public health measures during outbreaks and the guidance of vaccine clinical trials (by pharmaceutical industry colleagues; e.g. through facilitating selection of the study population).
This project will require innovative approaches for linking within-host viral dynamics models and population-scale transmission models, and aligns with the EPSRC “Tackling infections” and “Healthcare technologies” research themes. It falls under the EPSRC mathematical biology research area. Key mathematical approaches that will be used in the project include stochastic and deterministic transmission modelling, Bayesian parameter inference and numerical solution of ODEs, among other techniques. A key challenge when considering the association between viral infections and neurological conditions is that acute viral infection and the onset of neurological disease typically occur years apart; novel techniques are required to bridge this temporal gap using models. This project therefore involves designing multi-scale epidemiological modelling frameworks that can be used to understand dynamics over long timescales.
[Project chosen by Lewis Chinery, SABS R3 Programme 2020]
Supervisors
- Charlotte M Deane (Department of Statistics, University of Oxford)
- Dr Jeliazko (GSK)
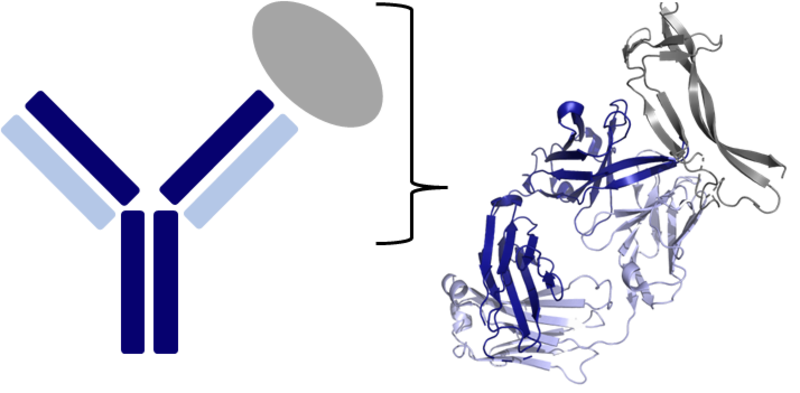
Cancer, rheumatoid arthritis, and many infectious diseases such as SARS-CoV-2 can all be treated, to an extent, using antibody drugs. These therapeutics are incredibly powerful given antibodies’ high binding affinity and specificity to their disease targets. However, developing such treatments often takes approximately ten years and over $1 billion in investment. Speeding up and lowering the cost of this process is therefore essential to the development of more affordable, effective treatments in the coming decades.
This research project was on developing novel computational advances to the initial ‘discovery’ stage of the drug development pipeline. Discovery can last approximately five years and involves testing and optimising tens of thousands of potential antibody-leads for their neutralising capability, developability, and immunogenicity.
Based within the Oxford Protein Informatics Group (OPIG) in the Department of Statistics, Lewis used advanced deep learning methods and other computational tools to enable researchers to move some of these in vitro discovery stage tests in silico, helping reduce development times and costs.
As one of many outputs from this project, Lewis created an antibody binding site (paratope) prediction tool – Paragraph (Chinery et al 2023. Bioinformatics). Paragraph leverages recent advances in graph neural networks to predict which amino acids of an antibody are likely to take part in binding its target. This knowledge is useful as different 3D orientations of an antibody-antigen complex can potentially lead to varying on- and off-target effects.
[Project chosen by Siting Miao, SABS R3 Programme 2020]
Supervisors
- Eamonn Gaffney (Maths)
- Helen Byrne (Maths)
- James Yates (GSK)
Interleukin-2 (IL-2) is a cytokine which can both promote and suppress immunity by binding to effector and regulatory T cells respectively. The seemingly contradictory effects of IL-2 have raised interest in selectively stimulating the effector or the regulatory T cells to promote or suppress immunity by generating different formulations of IL-2 that preferentially stimulate certain types of T cells. In the context of treatment of tumours or immune-deficiencies such as HIV, we want to preferentially stimulate effector T cells; while for autoimmune disease, we want to preferentially stimulate regulatory T cells. For the rational development of a range of immune-modulatory therapeutics, it is important to understand the role of IL-2 in mediating interactions between different types of T cells. We will use mathematical modelling to investigate this problem.
In the literature, mathematical models have been developed to study the role of IL-2 in mediating interactions between different types of T cells. All of these models do not account for the fact that when IL-2 binds onto the cell surface, the subsequent binding activities will be driven by 2D diffusion of the proteins on the cell surface. The modelling results from Sengers et al. indicate the importance of such surface diffusion. This motivates us to develop a model which takes into account such spatial aspects of the binding process. Also, the downstream subcellular signalling pathways have not been incorporated in detail in these models.
In order to achieve the goal of understanding the role of IL-2 in mediating interactions between different types of T cells, we need to consider:
(i) Individual cell scale: the dynamics of IL-2 binding to T-cell receptors,
(ii) Cell-cell interactions: how different types of T-cells compete for IL-2 and the feedback between binding and its expression level, and
(iii) Subcellular scale: how binding of IL-2 to receptors on T-cells leads to downstream subcellular signalling. We will use mathematical modelling to investigate each of these problems.
We may also use our model to explain other observations in the literature and a longer-term goal of the research will be systematic model integration. There is also the prospect that GSK may provide data that can be used in the project, though progress is not contingent on this.
We will look at various methods for model reduction in developing an asymptotically accurate reduced model, which may exploit multiple timescales such as the small timescale of diffusion on the cell surface compared to the timescale of interest or the large cell numbers in population studies. Numerical simulation will also be used to confirm the validity of these approximations in select, discerning examples.
We will also exploit sensitivity analysis to identify the core discriminants of the level of IL-2 signalling, both for an individual cell and a population of different cell types, as well as identifying potential rate limiting features. This will also help identify the potential for therapeutic targeting of IL-2 signalling.
Further, we will also use numerous tools from non-linear dynamical systems, such as bifurcation analysis, to identify any prospect for robust signalling that is insensitive to modulation, such as hysteresis, or what may drive auto-immune flares, such as excitability, and whether these complex behaviours may still be controlled.
[Project offered to students on the ILESLA Programme in 2025]
Supervisors
- Eamonn Gaffney (Maths)
- Tim Elliott (NDM)
- Brett Howell (GSK)
Activation of CD4⁺ T cells requires the presentation of short peptide fragments by MHC class II (MHC-II) molecules on the surface of antigen-presenting cells (APCs). This process is central to maintaining immune tolerance and coordinating T-cell responses, including within the tumour microenvironment. While the MHC-I pathway—which presents peptides derived from intracellular proteins to CD8⁺ T cells—is relatively well-characterised, the MHC-II pathway is less quantitatively understood. MHC-II presents peptides almost exclusively derived from extracellular proteins, which are first internalised and then transported through the compartments of the endosome-lysosome system, where they are degraded to peptides. Peptides are then loaded onto MHC-II molecules within this system via the action of HLA-DM, which exchanges a placeholder bound to MHC-II with high-affinity peptide. Once formed, the MHC-II peptide complex is trafficked to the cell surface and inserted into the cell plasma membrane for recognition by cognate CD4+ T cells to induce an immune response.
The goal of the full DPhil project is to develop a mechanistic computational model of the MHC-II antigen processing pathway, integrating protease specificity, HLA-DM editing, and MHC-II binding kinetics to predict which peptides are presented on the cell surface. The project will utilise public datasets and data from Professor Elliott’s laboratory. Models of the different subprocesses will employ both mechanism-based approaches (e.g., mass-action ordinary differential equations) and data-driven methods (e.g., deep learning). The outputs will be integrated into a predictive scoring system indicating the likelihood of peptide presentation for further experimental validation. Although beyond the immediate scope of this project, such models could ultimately inform the selection of epitopes for cancer vaccine design.
For the rotation project, the focus will be on modelling proteolytic digestion within endosomal-lysosomal compartments. Proteins are cleaved by proteases, and the pattern of cleavage is influenced by the protein sequence, structure, accessibility, and enzyme specificity. Mass spectrometry datasets provide high-resolution experimental measurements of cathepsin-generated peptides, which will be used to train deep learning models to predict proteolytic outcomes. This enables in silico simulation of antigen fragmentation, an important first step in identifying peptides potentially available for MHC-II binding.
The student can reasonably expect to have examined existing computational models of the proteasomal processing within the MHC-I pathway, gaining insight into an example of how data-driven approaches have been applied in antigen processing. The student should also be able to implement an initial iteration of a model for protein cleavage within the endosomal–lysosomal compartment for MHC-II peptide generation. This early model will serve as a prototype to enable a preliminary consideration of comparison to experimental data and validation strategies. However, explicit validation of the model’s predictions, as well as the iterative cycles of testing, refinement, and optimization necessary for a robust and generalizable model, will only be undertaken as part of the full DPhil project. In this context, the rotation project provides an essential opportunity to establish a prototype model, and outline potential validation approaches. This will form a foundation for more comprehensive model development during the long-term research program should the full DPhil project be pursued.
The BBSRC provides details of past projects in transformative technologies it has supported, their outcome and impacts, and other context including a review of their technology development portfolio.
